
This technique was first proposed by Dixon as the original two-point Dixon method, which acquires two sets of images at echo times when water and fat protons are in phase and out of phase. Fat–water decomposition MRI, one of the quantitative MRI techniques, is highly suitable for fat fraction quantification due to its ability to separate MRI signals from water and fat protons based on their chemical shift difference. Quantitative magnetic resonance imaging (MRI) provides an excellent approach for noninvasively measuring the fat composition in various organs, including muscle tissue. More importantly, the degree of intramuscular fat infiltration serves as a marker for disease severity and progression. Previous studies have shown that fat infiltration can be observed in thigh muscles due to diseases such as neuromuscular and metabolic disorders or age-related muscle atrophy (sarcopenia).
#Segmentation imuscle manual
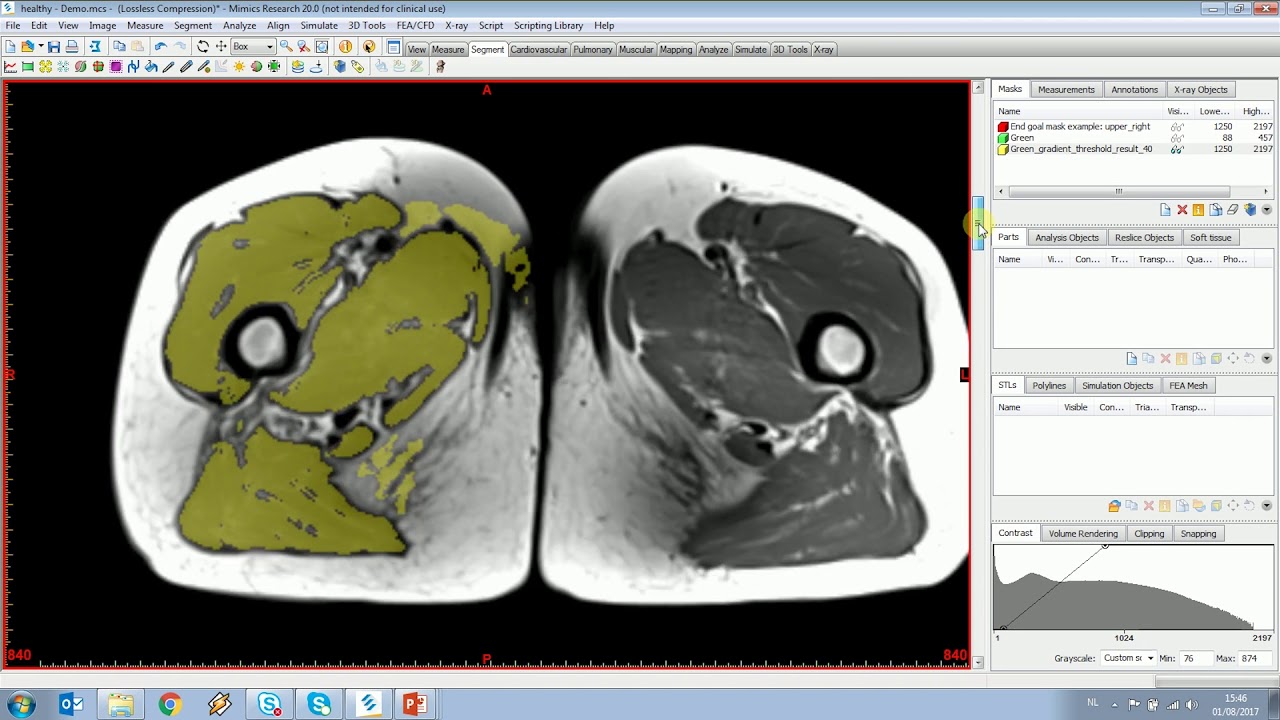
This automated thigh muscle segmentation exhibits excellent accuracy and higher reproducibility in fat fraction estimation compared to manual segmentation, which can be further used for quantifying fat infiltration in thigh muscles. A preliminary quantitative analysis was performed using two-sample t test to detect possible differences in meanFF between 14 normal and 14 abnormal (with fat infiltration) thighs in Dataset 2 using automated segmentation, and significantly higher meanFF was detected in abnormal thighs. The reproducibility in meanFF was calculated using intraclass correlation coefficients (ICCs) for the repeated scans, and automated segmentation produced overall higher ICCs than manual segmentation (0.921 vs. The average percent difference (absolute) in volume was 7.57%, and the average difference (absolute) in mean fat fraction (meanFF) was 0.17%. The average Dice coefficients between manual and automated segmentation were > 0.85. The segmentation accuracy was evaluated on an independent testing set (3 × 3 repeated scans in Dataset 1 and four scans in Dataset 2). A U-net was trained using 23 scans (16 from Dataset 1, seven from Dataset 2) to automatically segment four functional muscle groups: quadriceps femoris, sartorius, gracilis and hamstring. This study was performed using a public reference database (Dataset 1, 25 scans) and a local clinical dataset (Dataset 2, 21 scans). This study developed an automated whole thigh muscle segmentation method using deep learning for reproducible fat fraction quantification on fat–water decomposition MRI. This would open up various applications including personalization of biomechanical simulation and quantitative evaluation of muscle atrophy.Time-efficient and accurate whole volume thigh muscle segmentation is a major challenge in moving from qualitative assessment of thigh muscle MRI to more quantitative methods. The proposed method allows an accurate patient-specific analysis of individual muscle shapes in a clinical routine.
One application of the uncertainty metric in active-learning is demonstrated, and the proposed query pixel selection method considerably reduced the manual annotation cost for expanding the training data set. We evaluated validity of the uncertainty metric in the multi-class organ segmentation problem and demonstrated a correlation between the pixels with high uncertainty and the segmentation failure. These results were statistically significant improvements compared to the state-of-the-art hierarchical multi-atlas method which resulted in 0.845 ± 0.031 DC and 1.556 ± 0.444 mm ASD. The experiments showed a Dice coefficient (DC) of 0.891☐.016 (mean±std) and an average symmetric surface distance (ASD) of 0.994☐.230 mm over 19 muscles in the set of 20 CTs.
#Segmentation imuscle archive
We evaluated the performance of the proposed method using two data sets: 20 fully annotated CTs of the hip and thigh regions and 18 partially annotated CTs that are publicly available from The Cancer Imaging Archive (TCIA) database. The method uses Bayesian convolutional neural networks with the U-Net architecture, using Monte Carlo dropout that infers an uncertainty metric in addition to the segmentation label. We propose a method for automatic segmentation of individual muscles from a clinical CT.


 0 kommentar(er)
0 kommentar(er)
